I have recently started looking into ways to use Unreal Engine 4 for my visualizations and so far it looks quite promising. The data I usually work come from groundwater modeling simulations, therefore are not typical data one gets from modeling software such as Blender Houdini etc.
So the following is my way to get data from custom ascii files to UE4. For this guide I will assume that the data are organized as follows:
|
1 2 3 4 5 6 7 8 9 |
Nnodes Ntriangles X Y Z1 Z2 Z3 . . . ID1 ID2 ID3 . . . |
The first line is the number of nodes and the number of triangles. The procedure I’m following accepts only triangular data. so I have actually converted the quadrilaterals to triangles when I created the above file. Then Nnodes times is repeated the line that holds the x y coordinates. Note that in the file I provide 3 z coordinates. These correspond to ground surface, water table and bottom of the aquifer. in my case. Then I’m listing the ids of each of the Ntriangles of the mesh.
Inside UE4 I started from a C++ project and created an class named CustomMesh based on Actor class. The header file is the following
- Header file
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 |
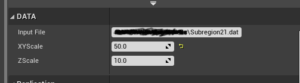
#pragma once #include "CoreMinimal.h" #include "GameFramework/Actor.h" #include "ProceduralMeshComponent.h" #include "CustomMesh.generated.h" UCLASS() class BUILING_ESCAPE_API ACustomMesh : public AActor { GENERATED_BODY() public: // Sets default values for this actor's properties ACustomMesh(); protected: // Called when the game starts or when spawned virtual void BeginPlay() override; virtual void OnConstruction(const FTransform& Transform) override; public: // Called every frame virtual void Tick(float DeltaTime) override; UPROPERTY(VisibleAnywhere, BlueprintReadWrite) UProceduralMeshComponent* PM; UPROPERTY(EditAnywhere, Category= "DATA") FString InputFile; UPROPERTY(EditAnywhere, Category= "DATA") float XYScale = 1000.f; UPROPERTY(EditAnywhere, Category= "DATA") float ZScale = 10.f; }; |
The most important line in the header file is the OnConstruction virtual method which is called every time the asset is transformed. Apart from the virtual method I defined 3 exposed properties InputFile, XYscale, Zscale. There is also a UProceduralMeshComponent that holds the actual data. This actually should not be exposed however I did it as I’m still experimenting with this. This header file will create a category DATA as shown below where the data can be updated inside the editor.
- Implementation file
- Constructor
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 |
#include "CustomMesh.h" #include "Misc/FileHelper.h" // Sets default values ACustomMesh::ACustomMesh() { // Set this actor to call Tick() every frame. You can turn this off to improve performance if you don't need it. PrimaryActorTick.bCanEverTick = true; PM = CreateDefaultSubobject<UProceduralMeshComponent>(TEXT("Procedural mesh")); SetRootComponent(PM); PM->bUseAsyncCooking = true; } |
In the constructor all I’m doing is to create the object and set it as root. Note also that I have included the Filehepler.h which is used for reading the custom file.
-
- OnConstruction
First we clear out and existing mesh in the object and initialize required variables.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 |
PM->ClearAllMeshSections(); TArray<FString> FileContent; FVector LowerCorner(999999999.f,999999999.f,999999999.f); FVector UpperCorner(-999999999.f,-999999999.f,-999999999.f); FVector Center(0.f, 0.f,0.f); int Nverts = 0; int Ntris = 0; int iline = 0; FVector T = Transform.GetLocation(); TArray<FVector> GSEVertices; TArray<FVector> BOTVertices; TArray<FVector> WTVertices; TArray<FVector> Normals; TArray<int32> Indices; TArray<FColor> ColorVert; |
Then I read the first line to get the number of vertices and number of triangles.
|
1 2 3 4 5 6 7 8 9 10 11 12 |
const bool bTF = FFileHelper::LoadANSITextFileToStrings(*InputFile,NULL, FileContent); if (bTF){ if (FileContent.Num() > 0){ //Read the number of vertices and triangles TArray<FString> Out; FileContent[iline].ParseIntoArray(Out, TEXT(" "), true); iline++; if (Out.Num() > 1){ Nverts = FCString::Atoi(*Out[0]); Ntris = FCString::Atoi(*Out[1]); } } |
Next we read the vertices. Note that while reading the x y coordinates we identify the bounding box of the data so that we can translate the mesh into the center of the transform. Otherwise the mesh will be located somewhere far away of the gizmo icon.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 |
if (FileContent.Num() > Nverts) { for (int i = 0; i < Nverts; ++i) { TArray<FString> Out; FileContent[iline].ParseIntoArray(Out, TEXT(" "), true); iline++; if (Out.Num() > 4) { float x = FCString::Atof(*Out[0])/XYScale + T.X; float y = FCString::Atof(*Out[1])/XYScale + T.Y; float t = FCString::Atof(*Out[2])/ZScale + T.Z; float w = FCString::Atof(*Out[3])/ZScale + T.Z; float b = FCString::Atof(*Out[4])/ZScale + T.Z; if (x > UpperCorner.X) UpperCorner.X = x; if (y > UpperCorner.Y) UpperCorner.Y = y; if (t > UpperCorner.Z) UpperCorner.Z = t; if (x < LowerCorner.X) LowerCorner.X = x; if (y < LowerCorner.Y) LowerCorner.Y = y; if (t < LowerCorner.Z) LowerCorner.Z = t; GSEVertices.Add(FVector(x,y,t)); Normals.Add(FVector::UpVector); BOTVertices.Add(FVector(x,y,b)); WTVertices.Add(FVector(x,y,w)); ColorVert.Add(FColor::MakeRandomColor()); } } Center = (UpperCorner + LowerCorner)/2; for (int i = 0; i < GSEVertices.Num(); ++i) { GSEVertices[i] = GSEVertices[i] - Center; BOTVertices[i] = BOTVertices[i] - Center; WTVertices[i] = WTVertices[i] - Center; } } |
Next we read the triangles. The triangles are stored as 1D vector.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 |
if (FileContent.Num() > Nverts + Ntris) { for (int i = 0; i < Ntris; ++i) { //UE_LOG(LogTemp, Warning, TEXT("%d"), i); TArray<FString> Out; FileContent[iline].ParseIntoArray(Out, TEXT(" "), true); iline++; if (Out.Num() > 2) { Indices.Add(FCString::Atoi(*Out[0])-1); Indices.Add(FCString::Atoi(*Out[2])-1); Indices.Add(FCString::Atoi(*Out[1])-1); } } } |
Last we create the custom meshes. For each Z coordinate I create one mesh, which set to visible after the creation.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 |
PM->CreateMeshSection_LinearColor(0, GSEVertices, Indices, TArray<FVector>(), TArray<FVector2D>(), TArray<FLinearColor>(), TArray<FProcMeshTangent>(),true); PM->CreateMeshSection_LinearColor(1, WTVertices, Indices, TArray<FVector>(), TArray<FVector2D>(), TArray<FLinearColor>(), TArray<FProcMeshTangent>(),true); PM->CreateMeshSection_LinearColor(2, BOTVertices, Indices, TArray<FVector>(), TArray<FVector2D>(), TArray<FLinearColor>(), TArray<FProcMeshTangent>(),true); /*PM->CreateMeshSection(0, Vertices, Indices, Normals, TArray<FVector2D>(), ColorVert, TArray<FProcMeshTangent>(), true);*/ PM->SetMeshSectionVisible(0,true); PM->SetMeshSectionVisible(1,true); PM->SetMeshSectionVisible(2,true); |
So far this is adequate to create the custom mesh. However created a blueprint class based on the Custom Mesh class which I dragged into my scene.
The above code does not read the UV coordinates. Soon I’ll add this info in the file and show some figures.
To be continued…